True Color (RGB)
FileSize: 139787
Format: JPEG
Width: 800
Height: 771
Depth: 8
StorageType: truecolor
NumberOfColors: 0
ResolutionUnit: inch
XResolution: 72.000000
YResolution: 72.000000
http://us.fun.marinov.net/store/Village%20Scenery/Village%20Scenery%20-%201.jpg

Indexed
FileSize: 28933
Format: GIF
Width: 500
Height: 344
Depth: 8
StorageType: indexed
NumberOfColors: 8
ResolutionUnit: centimeter
XResolution: 72.000000
YResolution: 72.000000
FileSize: 97685
Format: JPEG
Width: 450
Height: 342
Depth: 8
StorageType: indexed
NumberOfColors: 256
ResolutionUnit: inch
XResolution: 72.000000
YResolution: 72.00000
Format: JPEG
Width: 450
Height: 342
Depth: 8
StorageType: indexed
NumberOfColors: 256
ResolutionUnit: inch
XResolution: 72.000000
YResolution: 72.00000
http://gsc.nrcan.gc.ca/labs/ebeam/images/sem8.jpg
 Binary
BinaryFileSize: 38062
Format: BMP
Width: 775
Height: 380
Depth: 8
StorageType: indexed
NumberOfColors: 2
ResolutionUnit: centimeter
XResolution: 118.430000
YResolution: 118.430000
http://baec.tripod.com/AUGUST01/pics/binary_counter.bmp
************************************************
For the second part of the activity, a scanned image was enhanced using the threshold value from the histogram. Thresholding is basically making the region of interest (ROI) of the image well separated from the background. In this activity, a scanned image of a leaf was used.
 The image cropped using gimp. The colors were also inverted since I wanted the leaf to be the region of interest. Using scilab, the image was converted to grayscale. Its histogram and image properties were also obtained using the code below. The results then followed.
The image cropped using gimp. The colors were also inverted since I wanted the leaf to be the region of interest. Using scilab, the image was converted to grayscale. Its histogram and image properties were also obtained using the code below. The results then followed.
I=imread("C:\Documents and Settings\mimie\Desktop\leafginv.jpg");
I=im2gray(I);
//histogram
a = 1;
x= [];
y= [];
for i = 0:256
m = find(I== i);
x(a) = i;
y(a) = length(m);
a = a + 1;
end
scf(1)
plot(x,y/max(y)) //end of histogram routine
imshow(I)
imfinfo('C:\Documents and Settings\mimie\Desktop\leafginv.jpg')
 Image Properties:
Image Properties:
filename: C:\Documents and Settings\mimie\Desktop\leafginv.jpg
filesize:7359
format: jpeg
width: 207
height: 138
depth: 8
storage type: indexed
number of colors: 256
resolution unit: inch
x resolution: 0
y resolution: 0
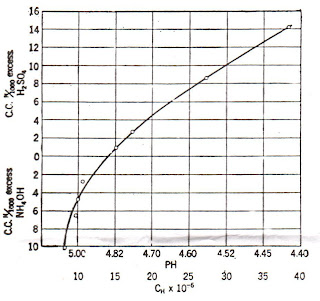
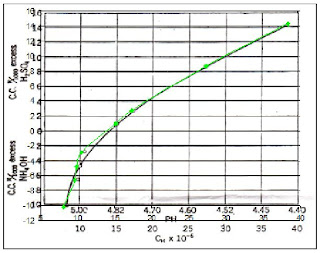
The histogram of a grayscale image runs from 0 to 255, 0 being black and 255 being white. From the histogram obtained, it can be seen that the ROI (around the second peak) is well separated from the background (the first peak). Also,the threshold value when this image is to be converted into binary is somewhere between 50/255 to 120/255. I used the average [(120+50)/2]/255 as the threshold value and obtained

Converting to binary image (in the command window):
Using the area calculation from 1, the area obtained using Green's theorem, by pixel counting, and the relative error between the two are:
area = 15979
theo = 15978
%error: 0.0062586
To verify the threshold values obtained using scilab, Gimp can be used.
1. Click 'Colors' -->'Threshold' (This will turn the grayscale image into black and white.)
2. Adjust the arrows (or the values) until the ROI is well separated from the background
3. The threshold value for the image is just the minimum/maximum.
Acknowledgemets to Jeric Tugaff for the histogram code (posted in Ma'am Jing's blog) and Ma'am Jing for her suggestions.
I give myself a 9/10 for this activity.
Format: BMP
Width: 775
Height: 380
Depth: 8
StorageType: indexed
NumberOfColors: 2
ResolutionUnit: centimeter
XResolution: 118.430000
YResolution: 118.430000
http://baec.tripod.com/AUGUST01/pics/binary_counter.bmp
************************************************
For the second part of the activity, a scanned image was enhanced using the threshold value from the histogram. Thresholding is basically making the region of interest (ROI) of the image well separated from the background. In this activity, a scanned image of a leaf was used.
 The image cropped using gimp. The colors were also inverted since I wanted the leaf to be the region of interest. Using scilab, the image was converted to grayscale. Its histogram and image properties were also obtained using the code below. The results then followed.
The image cropped using gimp. The colors were also inverted since I wanted the leaf to be the region of interest. Using scilab, the image was converted to grayscale. Its histogram and image properties were also obtained using the code below. The results then followed.I=imread("C:\Documents and Settings\mimie\Desktop\leafginv.jpg");
I=im2gray(I);
//histogram
a = 1;
x= [];
y= [];
for i = 0:256
m = find(I== i);
x(a) = i;
y(a) = length(m);
a = a + 1;
end
scf(1)
plot(x,y/max(y)) //end of histogram routine
imshow(I)
imfinfo('C:\Documents and Settings\mimie\Desktop\leafginv.jpg')
 Image Properties:
Image Properties:filename: C:\Documents and Settings\mimie\Desktop\leafginv.jpg
filesize:7359
format: jpeg
width: 207
height: 138
depth: 8
storage type: indexed
number of colors: 256
resolution unit: inch
x resolution: 0
y resolution: 0
The histogram of a grayscale image runs from 0 to 255, 0 being black and 255 being white. From the histogram obtained, it can be seen that the ROI (around the second peak) is well separated from the background (the first peak). Also,the threshold value when this image is to be converted into binary is somewhere between 50/255 to 120/255. I used the average [(120+50)/2]/255 as the threshold value and obtained

Converting to binary image (in the command window):
i=im2bw(I,85/255);
imshow(i)
imshow(i)
Using the area calculation from 1, the area obtained using Green's theorem, by pixel counting, and the relative error between the two are:
area = 15979
theo = 15978
%error: 0.0062586
To verify the threshold values obtained using scilab, Gimp can be used.
1. Click 'Colors' -->'Threshold' (This will turn the grayscale image into black and white.)
2. Adjust the arrows (or the values) until the ROI is well separated from the background
3. The threshold value for the image is just the minimum/maximum.
Acknowledgemets to Jeric Tugaff for the histogram code (posted in Ma'am Jing's blog) and Ma'am Jing for her suggestions.
I give myself a 9/10 for this activity.